Integrated Segmentation And Interpolation
We propose an integrated segmentation and interpolation framework, which can handle 3D and 4D sparse data based on a novel level set scheme. In this new framework, the level set implicit function is interpolated by radial basis functions (RBFs), and its interface can propagate in a sparse volume using data information where available and RBF-based interpolation in the gaps. The interpolation process uses segmentation information rather than pixel intensities for increased robustness and accuracy. The method supports any spatial configurations of sets of 2D slices having arbitrary positions and orientations.
The proposed method is validated quantitatively and/or subjectively on artificial and real data including MRI and CT scans and is compared with the traditional sequential approach, which interpolates the images first using a state-of-the-art image interpolation method, and then segments the interpolated volume in 3D or 4D. Thanks to the interdependency of the segmentation and interpolation processes, the integrated framework yields similar segmentation results to the sequential approach but provides a more robust and accurate interpolation. In particular, the interpolation is more satisfactory in cases of large gaps, due to the method taking into account the global shape of the object, and it recovers better topologies at the extremities of the shapes where the objects disappear from the image slices. As a result, the complete integrated framework provides more satisfactory shape reconstructions than the sequential approach.
 |
| Examples of sparse data sets. (Left) simple stack of parallel slices, (Middle) standard spatial configuration of a cardiac MRI: stack of parallel short-axis slices plus a few long-axis ones, and (Right) radial dataset. |
Proposed framework
The proposed integrated segmentation and interpolation framework is based on a novel RBF-interpolated level set method. This method can handle sets of 2D slices having any spatial configuration, i.e. made of any number of slices having arbitrary positions and orientations.
- Robustness to noise
 |  |  |
| (a) | (b) | (c) |
 |  |  |
| (d) | (e) | (f) |
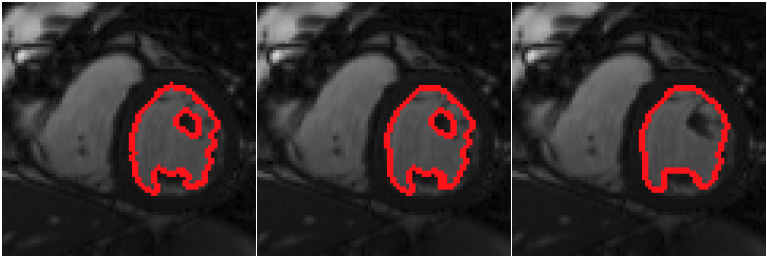
Robust to noise property of the proposed framework. (a) initial noisy image, segmentation by the (b) narrow-band level set with piecewise constant model, (c) Chan-Vese level set scheme, and (d) proposed method; (e) initial noisy image with 'slices' of missing information and (f) segmentation by the proposed method with gaps being interpolated.
- Preserving small objects or holes
 |
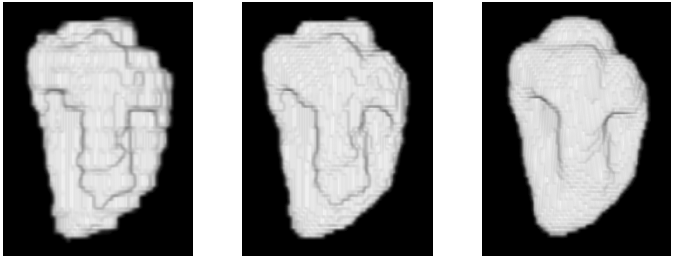 |
| Small object or hole preserving property of the proposed framework - the modeled object is the cavity of the left ventricle (LV) of a heart from short-axis MRI scan. (Left column) initial segmentation and locally interpolated surface using a sharp RBF, (middle column) final segmentation and globally interpolated surface using a flat RBF, and (right column) processing of the dataset using a flat RBF only without preliminary segmentation. The small object or hole preserving approach (first two columns) segments the papillary muscle inside the LV cavity, while the simplest approach with a single flat RBF in the right column treats it as noise. |
Example results - 3D artificial data
| Our Previous Method [MIUA'10] | 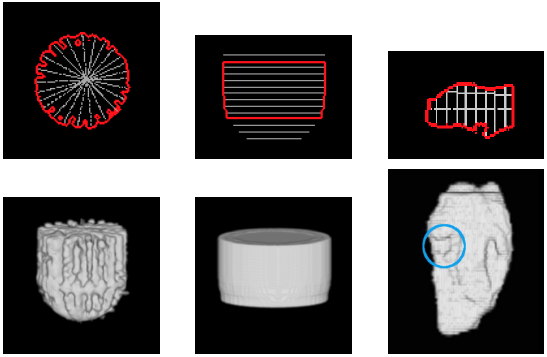 |
| Proposed Method | 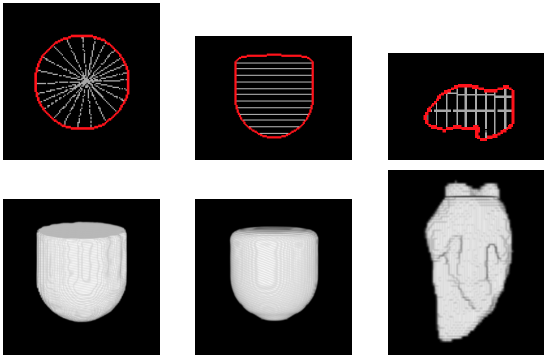 |
| 3D segmentation and interpolation of artificial data. (Left column) radial dataset – top: central horizontal slice, bottom: 3D view, (middle column) axial dataset – top: central vertical slice, bottom: 3D view, and (right column) LV cavity model – top: central vertical slice, bottom: 3D view. The blue circle highlights a failure of our previous method [MIUA'10] to recover the topology of part of the LV cavity. |
Example results - 3D real data
| Low-resolution phantom | High-resolution phantom | Kidney | Left ventricle | Acetabulum | |
| Sequential Method |  |  | 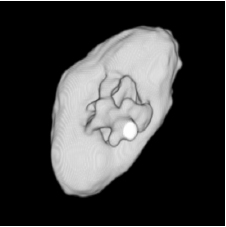 | 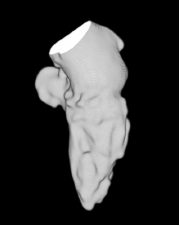 | 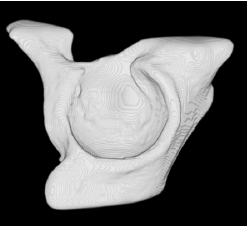 |
| Proposed Method |  |  | 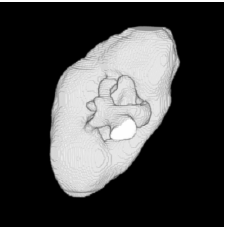 | 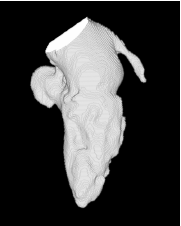 | 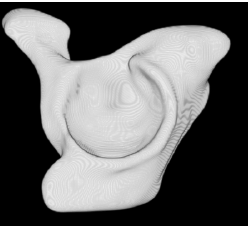 |
| Proposed Method + Vertical Slice |  |  | 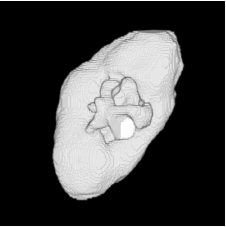 | 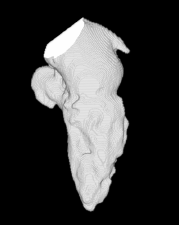 | 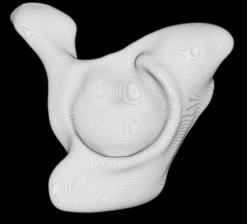 |
| Sequential Method | 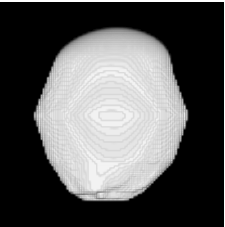 |  | 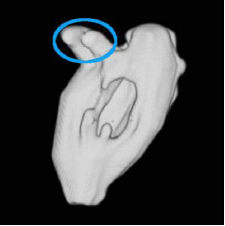 | 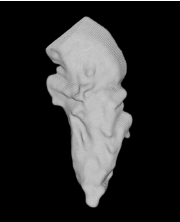 | 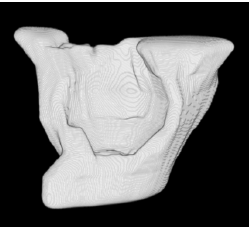 |
| Proposed Method |  |  | 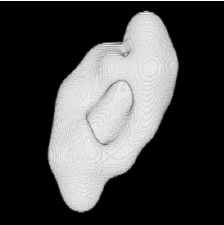 | 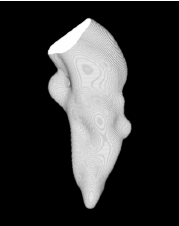 | 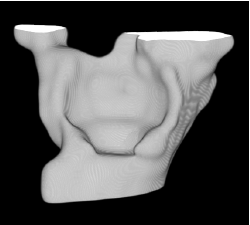 |
| Proposed Method + Vertical Slice |  |  | 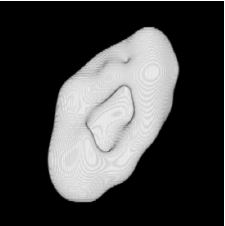 | 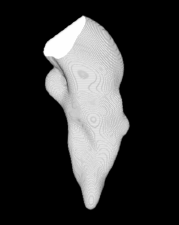 | 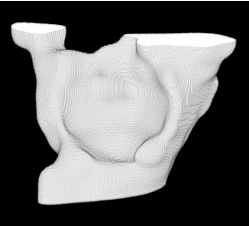 |
3D segmentation and interpolation of various shapes from real data at two different slice spacings (top: 5 pixels; bottom: 20 pixels), and from a variety of modalities. The blue circle highlights how sequential method struggles to reconstruct extremities of objects for large slice spacings, especially where a background of similar intensity can be confused with the object.
Example results - 3D real MRIs of brain ventricles
| Initial slices | T1 central slice | T2 central slice | Axial 3D view | Sagittal 3D view | |
| Our Previous Method [MIUA'10] | 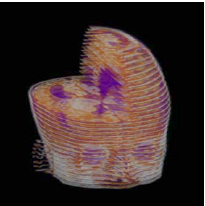 | 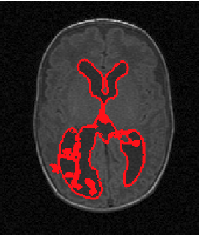 | 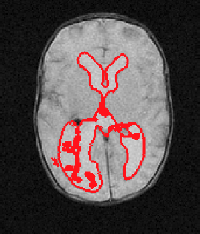 | 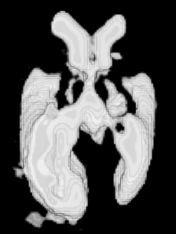 | 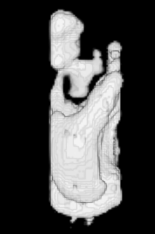 |
| Proposed Method |  | 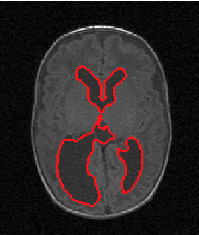 | 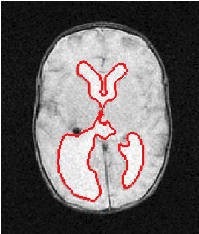 | 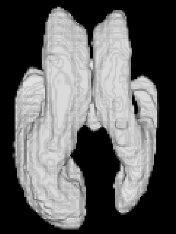 | 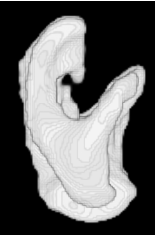 |
3D segmentation and interpolation of the ventricles of the neonatal brain from combined T1- and T2-weighted MRI scans. (Initial slices: top left quadrant removed for better visualisation of the ventricles in purple; red color: segmentation.)
Example results - 3D real cardiac MRIs of LV cavity
| Our previous method [MIUA'10] | Sequential method | Proposed method |
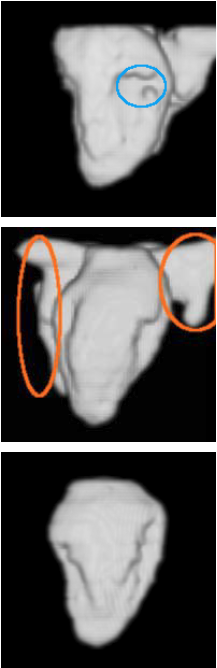 |  |  |
3D segmentation and interpolation of the LV cavity from three different cardiac MRI scans. The extraneous regions protruding from some LV cavities for all methods in some images (e.g. orange circles in the second row) are parts of the right ventricle which have been wrongly segmented by the chosen segmentation algorithm. The blue circles highlight situations where the proposed method recovered a better topology than our previous method and the sequential method.
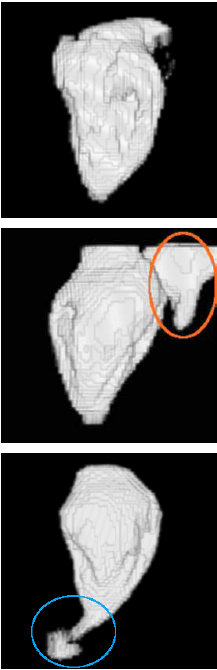
Example results - 4D real data
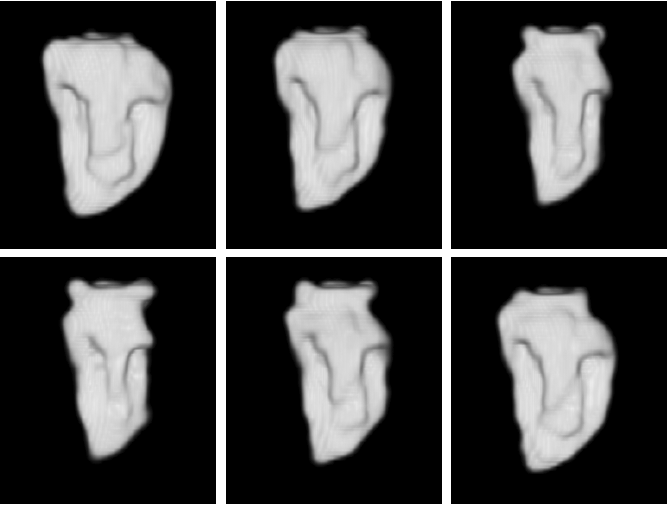 |
| 4D segmentation and interpolation of the LV cavity by the proposed method from a cardiac MRI (6 different time frames). Our previous method and the sequential method cannot handle 4D data. |
Example results - multiple regions
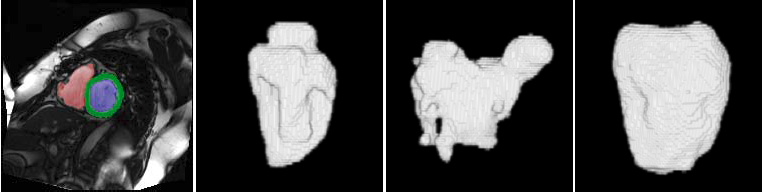 |
| Segmentation of multiple regions by the proposed method. From left to right: short-axis slice with the LV, RV and myocardium segmentations colored in blue, red, and green respectively, 3D reconstructions of the LV, the RV, and the myocardium. |
Example results - segmentation and interpolation in action
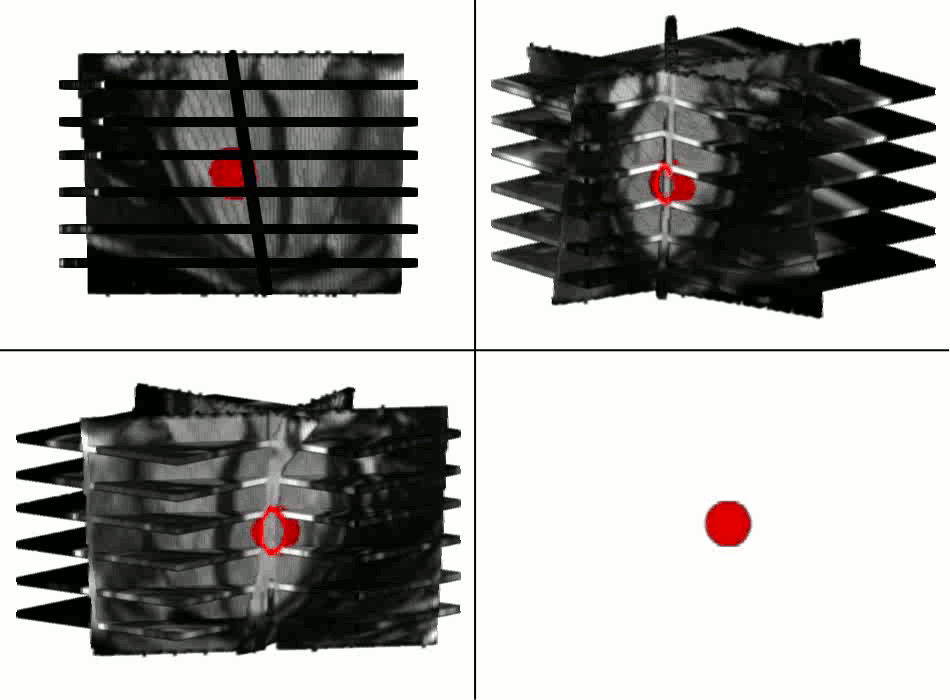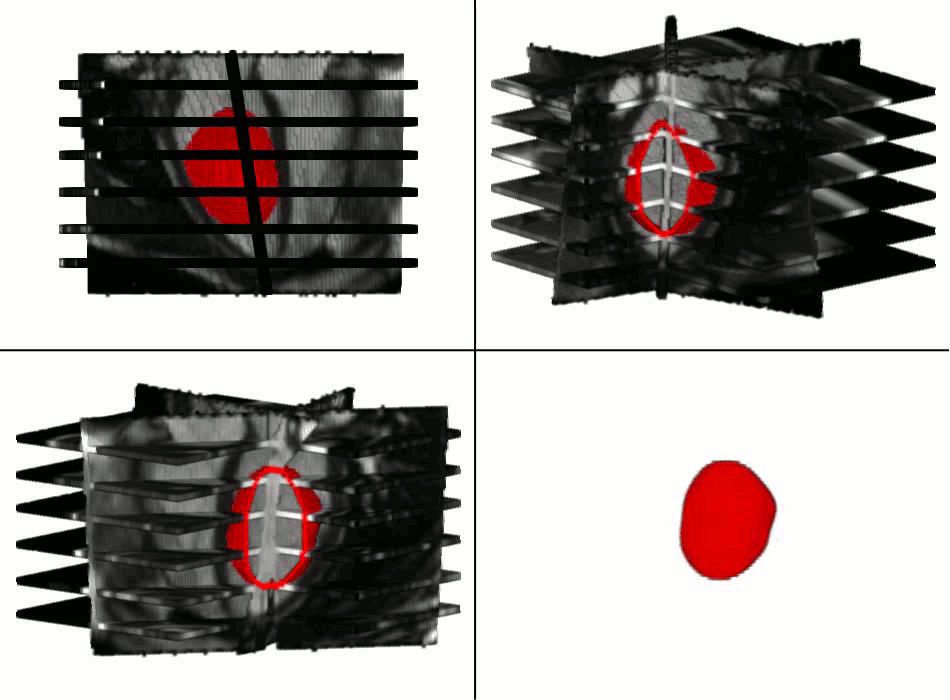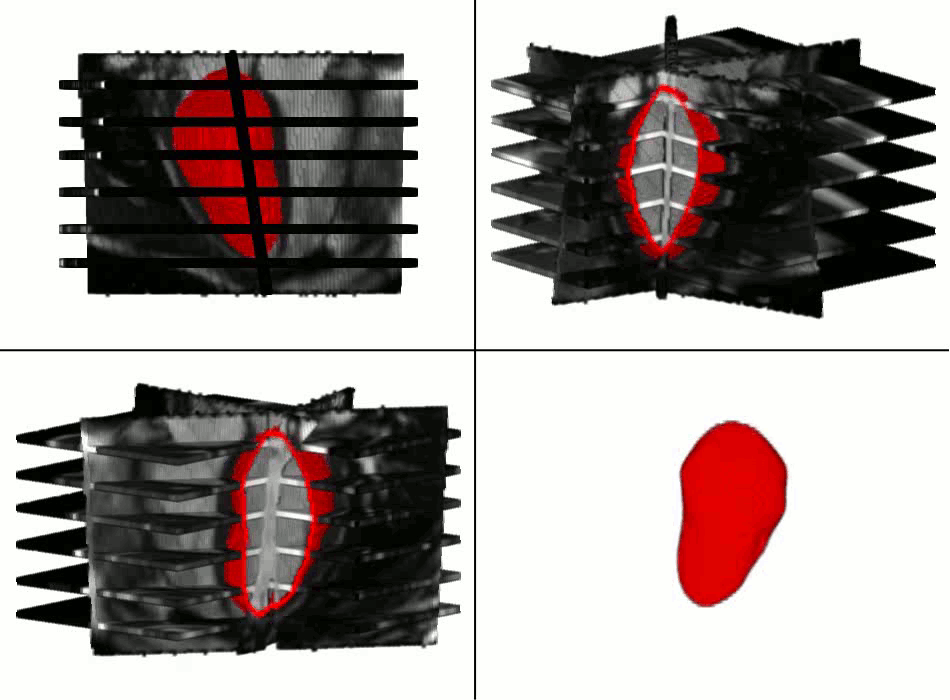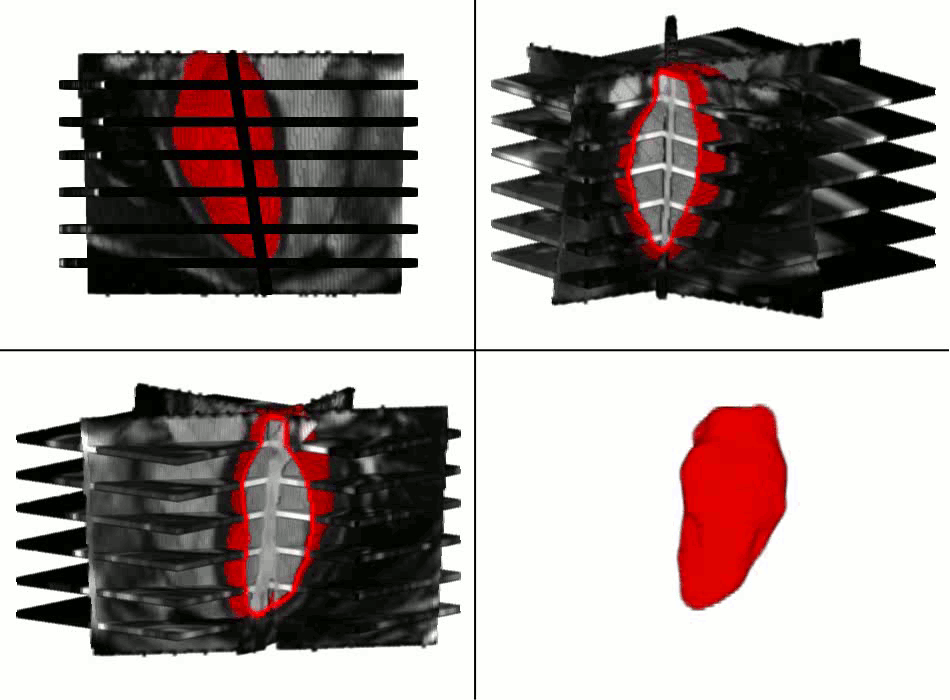 |
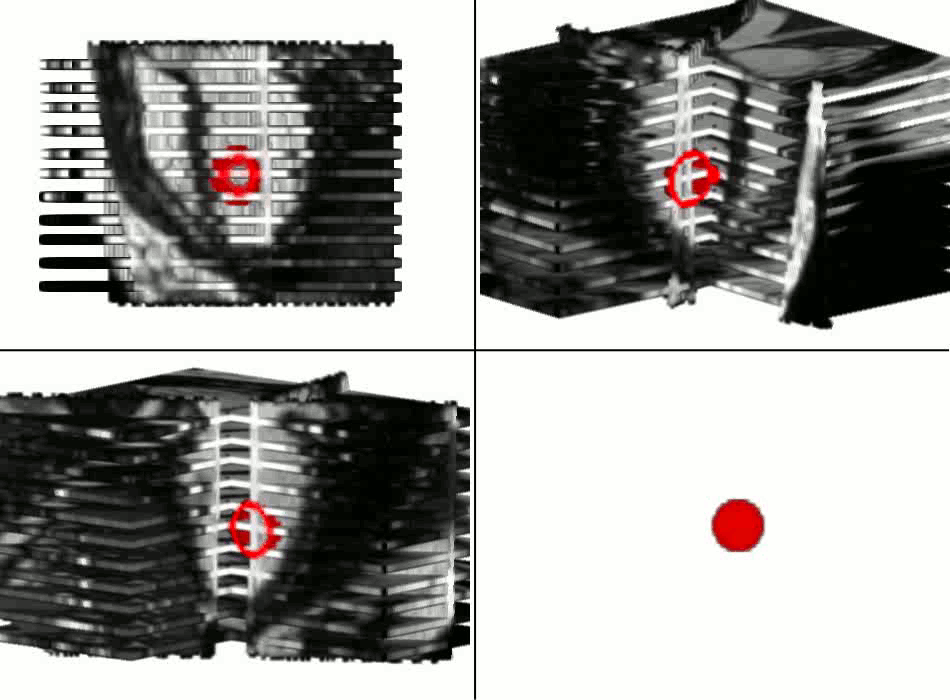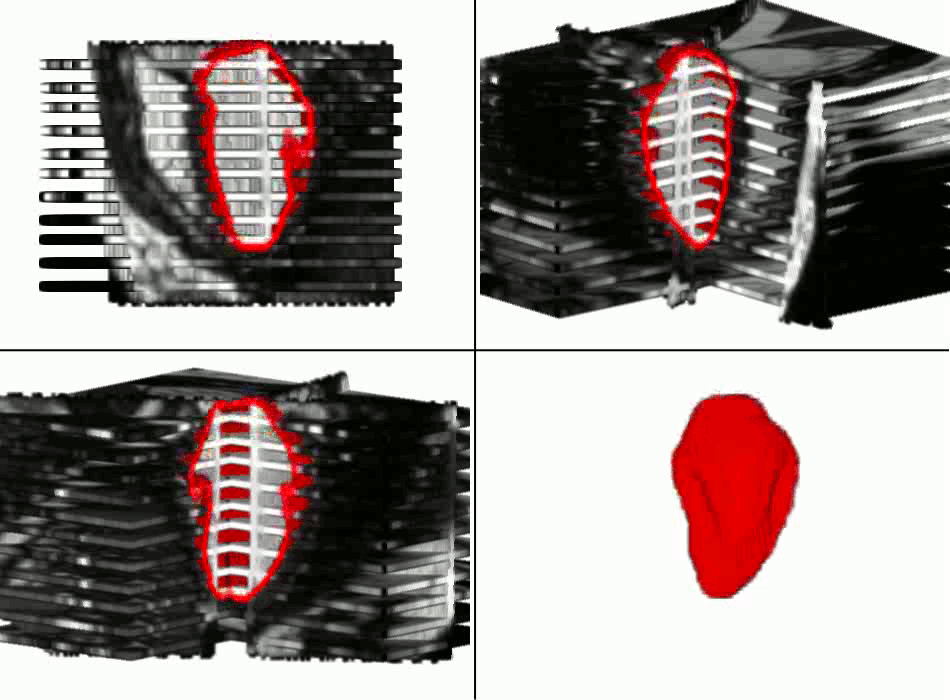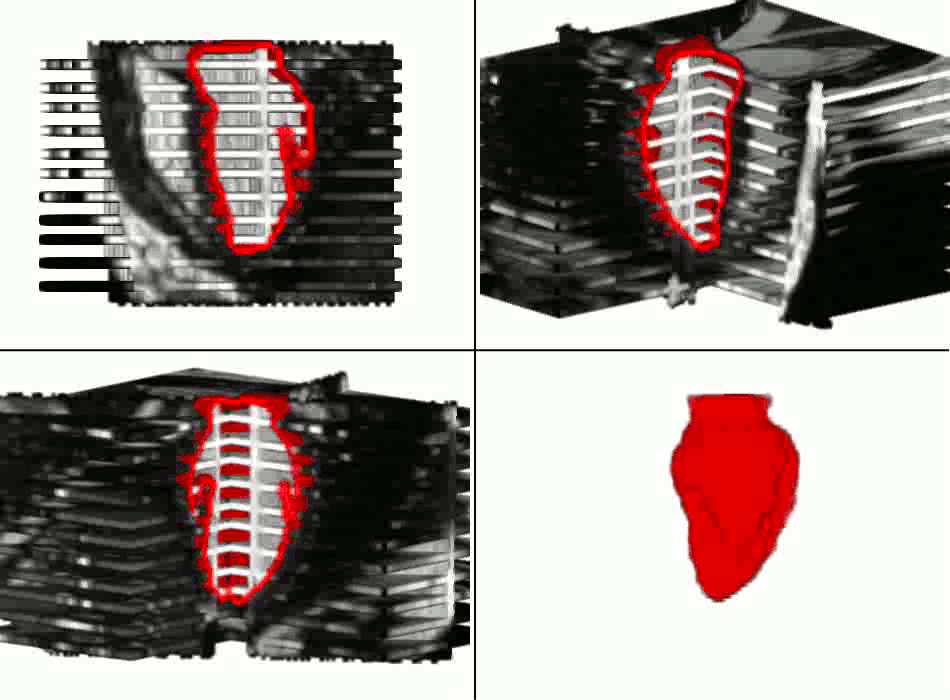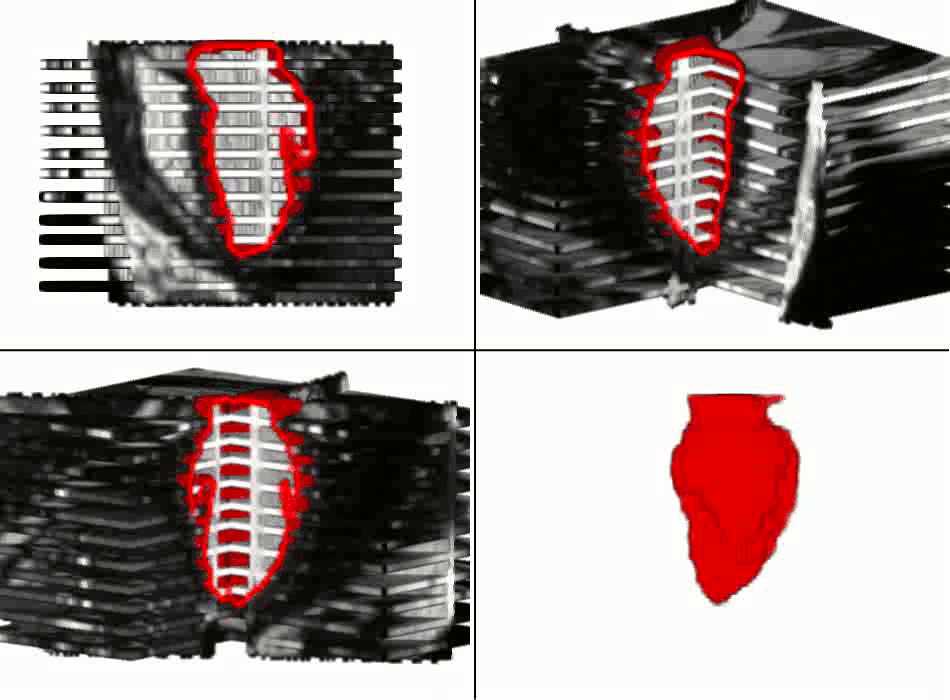 |
Publications
- A. Paiement, M. Mirmehdi, X. Xie, and M. Hamilton, Integrated Segmentation and Interpolation of Sparse Data, IEEE Transactions on Image Processing (T-IP), volume 23, issue 1, pages 110-125, January 2014.
- A. Paiement, M. Mirmehdi, X. Xie, and M. Hamilton, Simultaneous level set interpolation and segmentation of short- and long-axis MRI, In Proceedings of Medical Image Understanding and Analysis (MIUA), July 2010.
